Bioinformaticians handle an increasingly large and diverse set of tools and data. Meanwhile, researchers demand ever more powerful and convenient means to organise, find, compare, select, reuse and connect the available resources. These tasks often rely on consistent, machine-understandable descriptions of the underlying components, but these have been generally lacking in documentation and metadata developed ad hoc. There is therefore an urgent need for an ontology that unifies semantically the bioinformatics concepts in common use and, for the annotator, provides a comprehensive controlled vocabulary that is broadly applicable.
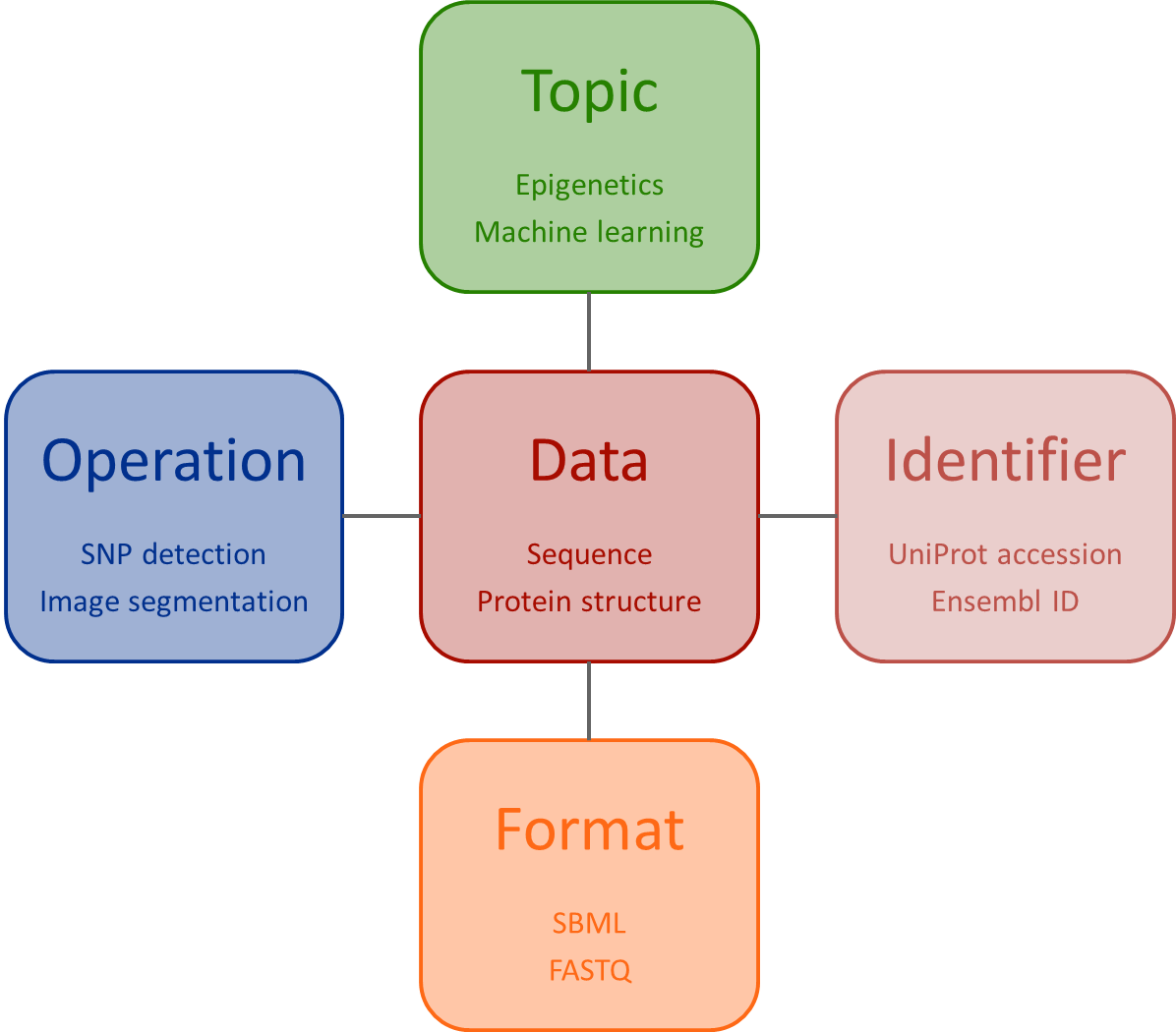
EDAM (EMBRACE Data and Methods) is an ontology of common bioinformatics operations, topics, types of data including identifiers, and formats. EDAM comprises common concepts (shared within the bioinformatics community) that apply to semantic annotation of resources such as:
As a rule (with a few exceptions) EDAM only includes concepts strictly in domain of bioinformatics/computational biology. General biology, medicine, or informatics concepts are (typically) not included in the EDAM Operation, Data, or Format sub-ontologies. However, broader inter-disciplinary topics are included in the EDAM Topic sub-ontology.
For example within drug design, the concepts related to sequence, structure, and molecular interactions analyses are part of EDAM, but ones related to pre-clinical and clinical trials are not. As another example, text mining in patient records and statistical analysis with respect to knowledge about diseases are in scope, but management of patient records or statistical analysis with respect to economy or administration are not.
EDAM includes 4 main sub-ontologies of concepts organised into simple hierarchies:
Noteworthy within the Data sub-ontology is:
These provide different semantic 'axes' for annotation. For example, annotation of a Web service might include:
EDAM has 4 components:
Concepts - These are well established and familiar bioinformatics concepts. Concepts have a name (term), a definition and one or more simple relations to other concepts defined in EDAM. Each concept has one or more intrinsic properties (reflected in the definition and relations).
Hierarchy - Every concept (excluding top-level concepts) is related to (typically) one other concept within the same sub-ontology by an is a (generalisation) relation. These relations define the sub-ontology hierarchies. All "child" concepts must share the intrinsic property of their "parent", in addition to having their own intrinsic properties.
Relations - Concepts are related by defined relation types, but these types of relation apply also between other entities outside of EDAM (for example artifacts semantically annotated by EDAM or another ontology)
Rules - There are simple rules dictating how different types of concepts are related within EDAM. They define which relations may be specified for which concepts. They reflect well established or self-evident principles.

Figure 1. The EDAM architecture is intentionally simple. Boxes indicate top-level concepts (sub-ontologies), and lines indicate types of relations that are maintained between concepts in EDAM.
Locations for download in OWL format:
http://edamontology.org/EDAM.owl (Always points to the last stable version)
Locations for download in OBO format:
http://edamontology.org/EDAM.obo (Always points to the last stable version. OBO-format version lacks certain details.)
All versions:
http://sourceforge.net/projects/edamontology/files/
Version 1.2 of EDAM has been released . It uses the concepts, relations and rules below. Contributions and suggestions are welcome.
EDAM is being actively developed:
For further information see the EDAM Wiki:
https://sourceforge.net/apps/mediawiki/edamontology/index.php?title=Main_Page
See the EDAM presentation at the BioOntologies SIG, ISMB 2011:
http://bio-ontologies.knowledgeblog.org/224
EDAM is available in the following Web-based ontology browsers:
EDAM is made available for everyone to use, with the following constraints on its use or redistribution (including online access):
The intellectual content of EDAM or of any of its parts cannot be included in other projects and artifacts unless agreed with the authors of EDAM.
If you use EDAM or its parts, please reference:
Ison, J., Kalaš, M., Jonassen, I., Bolser, D., Uludag, M., McWilliam, H., Malone, J., Lopez, R., Pettifer, S. and Rice, P. (2013). EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats. Bioinformatics, 29, 1325-1332.
doi: 10.1093/bioinformatics/btt113 PMID: 23479348
This article is freely available (Open Access).
All enquiries to Jon Ison (jison@ebi.ac.uk) CC'ing Matúš Kalaš (matus.kalas@uib.no)
Thanks for valuable discussions and contributions to Peter Rice, Inge Jonassen, Dan Bolser, Rodrigo Lopez, Gert Vriend, James Malone, Steve Pettifer, Hamish McWilliam, Alan Bleasby, Mahmut Uludag, László Kaján and others.
Feel free to subscribe to the mailing lists:
Once subscribed, you can mail the user and developer lists:
edamontology-announce is for announcements (very minimal traffic!) edamontology-developers is for technical discussions between EDAM developers / contributors. edamontology-users is for general discussions and announcements.
"A function or process performed by a tool; what is done, but not (typically) how or in what context."
e.g. "Sequence alignment", "Pairwise sequence alignment", "Sequence database search".
"Operation" concepts provide mostly fine-grained concepts for annotation of tool functions.
The top-level concepts are:
The top-level operations are necessarily coarse-grained (abstract) providing a navigable top-level. They serve as placeholders for other, more specific concepts lower down in the tree.
"A type of data in common use in bioinformatics."
e.g. "Sequence alignment", "Comparison matrix", "Phylogenetic tree" etc.
Data concepts:
The top-level concepts are:
Their meaning is:
Concepts within "Core data" are:
"A general bioinformatics subject or category, such as a field of study, data, processing, analysis or technology."
e.g. "Sequence analysis", "Alignment", "Sequencing", "Microarrays".
"Topic" concepts provide coarse-grained categories for annotation of diverse bioinformatics resources. They do not cover biology or computer science exhaustively.
The top-level concepts are:
"A specific layout for encoding a specific type of data in a computer file or memory."
e.g. "FASTA format", "PDB format", "mmCIF" etc.
"Format" concepts:
The top-level concepts are:
All concepts are nested under "Binary format", "Textual format" and "XML", with exception of pure "HTML" or "RDF" (and "BioPAX"). The "Format (typed)" branch arranges formats by type of data and provides an additional axis over (the same set of) concepts under "Binary format", "Textual format" and "XML".
"A label that identifies (typically uniquely) something such as data, a resource or a biological entity."
e.g. "UniProt accession", "EC number", "Gene symbol" etc.
"Identifier" concepts:
The top-level concepts are:
As for "Format", the "Identifier (typed)" branch provides an additional axis over (the same set of) concepts under "Accession" and "Name".
Defines a concept as a specialisation of another concept. If A is a B, then A is a specialisation of B, and B is a generalisation of A.
The is a relation is transitive: if A is a B and B is a C then A is a C.
All relations are transitive over is a: e.g. if A has input B and B is a C then A has input C, and if A is a B and B has input C then A has input C.
e.g. "Pairwise sequence alignment" is a "Sequence alignment"
Defines an "Operation" concept as reading (inputting) a "Data" concept.
e.g. "Sequence alignment construction" has input "Sequence"
Defines an "Operation" concept as writing (outputting) a "Data" concept.
e.g. "Sequence alignment construction" has output "Sequence alignment"
Defines a "Data" or "Operation" concept as being within the scope of a "Topic" concept.
e.g. "PolyA signal identification" has topic "Nucleic acid sequence analysis"
Defines that an "Identifier" concept identifies a "Data" concept.
e.g. "Sequence accession number" is identifier of "Sequence"
Defines that a "Format" concept is the format of a "Data" concept.
e.g. "Sequence format" is format of "Sequence record"
Rules define how concepts are related.
... a specialisation of a topic.
... a specialisation of an operation.
... inputs a type of data.
... outputs a type of data.
... within a topic.
... a specialisation of a type of data.
... within a topic.
... a specialisation of a data format.
... a format specification of a datatype.
... identifier of a datatype.
Various resources were analysed while constructing EDAM and were used as sources listing common bioinformatics concepts in scope.
Annotators may email Jon Ison (jison@ebi.ac.uk) and Matúš Kalaš (matus.kalas@bccs.uib.no) for help.
The expectation is for EDAM to be used alongside other ontologies for annotation where possible and desirable. For example, an operation that predicts specific features of a molecular sequence could be annotated with concepts from SO (Sequence Ontology) for the features.
If you have many annotations to do, it will help to familiarise yourself with EDAM first using a browser (see Viewing).
A Web service is considered as an arbitrary (but usually related) set of one or more operations, reducing the problem of Web service interoperation to one of compatibility between operations.
Operation
Input
Output
XML elements
Annotation of a WSDL file or associated XSD schema is possible at several levels. Assuming SAWSDL annotation (http://www.w3.org/TR/sawsdl/), the XML elements that may be annotated by EDAM concepts are:
NB. The input and output parameters should be annotated inside the XML Schema that defines them. In case of services that are not following the highly recommended document/literal wrapped SOAP-binding style, the <wsdl:part> inside <wsdl:message> can be annotated (the same applies to faults, but meanings of faults are not modelled by EDAM)
The following annotations might be useful but are not directly recommended by SAWSDL:
For details of incorporating the SAWSDL annotations into WSDLs and XSDs, see EDAM URIs and SAWSDL annotation.
SAWSDL mandates the use of sawsdl:modelReference attributes for annotation. The format of EDAM URIs used inside this attribute includes the ontology name (http://edamontology.org), main sub-ontology, and the unique identifier (ID) of the particular concept:
<xs:element name="elementName" sawsdl:modelReference="http://edamontology.org/subontology_id">
Where ...
The value of the sawsdl:modelReference attribute is a URI pointing to the concept definition. The URI to use is in case of EDAM includes the concept's sub-ontology:
So for these 3 concepts:
EDAM_topic:0182 EDAM_operation:0292 EDAM_data:0863
We'd have
http://edamontology.org/topic_0182 http://edamontology.org/operation_0292 http://edamontology.org/data_0863
Which can be used in SAWSDL annotation, e.g.
<wsdl:portType name="myService" sawsdl:modelReference="http://edamontology.org/topic_0182"> <sawsdl:attrExtension sawsdl:modelReference="http://edamontology.org/operation_0292> <xs:element name="outfile" sawsdl:modelReference="http://edamontology.org/data_0863>
If more than one annotation of an element is required, these can be given in the sawsdl:modelReference attribute delimited by space characters:
<wsdl:portType name="myService" sawsdl:modelReference="http://edamontology.org/topic_0182 http://edamontology.org/operation_0292">
NB. Such multiple annotations need not be in the same namespace, and need not at all to refer to the same ontology.
One peculiarity of the SAWSDL specification is that annotations on <wsdl:operation> element inside <wsdl:portType> should be handled using a <sawsdl:attrExtensions> element. This is not a requirement for other elements.
Importantly, the <sawsdl:attrExtension> element inside the wsdl:operation must be before <wsdl:input>, <wsdl:output> and <wsdl:fault> elements (so typically after the <wsdl:documentation> element).
For example:
<wsdl:portType name="Clustalw2PortType" sawsdl:modelReference="http://edamontology.org/topic_0186 http://edamontology.org/operation_0496">
<wsdl:operation name="submitClustalw2">
<wsdl:documentation>Submit a sequence and get a jobID</wsdl:documentation>
<sawsdl:attrExtensions sawsdl:modelReference="http://edamontology.org/operation_0496"/>
<wsdl:input message="submitClustalw2Msg"/>
<wsdl:output message="submitClustalw2ResponseMsg"/>
</wsdl:operation>
Some WSDL/XSD validators or SOAP libraries do not check for it, but some do require the strict order of these elements.
EMBOSS applications have been annotated using EDAM and these annotations appear in corresponding Web services.
Annotated WSDL files (and associated XSD data schema) are available from:
You will see a list of service end-points with WSDL URLs. For example:
To see the data schema associated with a WSDL, you must replace "?wsdl" with "?xsd=1", "?xsd=2" or "?xsd=3". For example:
The BioXSD XML schema (XSD) defines exchange formats of everyday bioinformatics data types. BioXSD aims to serve as the common, canonical data model for bioinformatics Web services. It includes commonly used types including sequences, sequence annotations, alignments and references to resources:
BioXSD has been annotated with EDAM concepts.
A catalogue of data resources (DRCAT) is being compiled as part of the EMBOSS project. Each entry in DRCAT gives metadata on a data resource available on the Web. The metadata includes "Query" lines describe the type(s) of data available, the data format, data identifier (used to query) and a URL from which data can be retrieved. The "Query" lines and the resources themselves are annotated with EDAM concepts.
A typical entry is shown below:
(NB. The format of EDAM ids has not been upgraded to version 1.0 yet. Will be done asap.)
ID PDB Acc DB-0070 Name The RCSB Protein Data Bank Desc A repository for 3D biological macromolecular structure data. URL http://www.rcsb.org/pdb/ Cat 3D structure databases EDAMres EDAM:0000693 | Tertiary structure EDAMdat EDAM:0000883 | Tertiary structure EDAMdat EDAM:0002085 | Structure annotation EDAMfmt EDAM:0001476 | pdb EDAMfmt EDAM:0001478 | pdbml EDAMfmt EDAM:0001477 | mmCIF EDAMfmt EDAM:0002331 | HTML EDAMid EDAM:0001127 | PDB ID Xref SP_explicit | None Xref SP_FT | None Xref EMBL_explicit | None Query EDAM:0002085 | EDAM:0002331 | EDAM:0001127 | http://www.pdb.org/pdb/explore/explore.do?structureId=%s Query EDAM:0000693 | EDAM:0001476 | EDAM:0001127 | http://www.pdb.org/pdb/files/%s.pdb Query EDAM:0000693 | EDAM:0001477 | EDAM:0001127 | http://www.pdb.org/pdb/files/%s.cif Query EDAM:0000693 | EDAM:0001478 | EDAM:0001127 | http://www.pdb.org/pdb/files/%s.xml Example EDAM:0001127 | 1rbp Email deposit@deposit.rcsb.org CCmisc EMBL DR line example "1OSN", /dbxref="PDB:12GS" Status Referenced
DRCAT development will proceed in harmony with bioDBCore, which proposes a community-defined, uniform, generic description of the core attributes of biological databases:
bioDBCore is under the auspices of the International Society for Biocuration:
All enquiries to Jon Ison (jison@ebi.ac.uk)
Bio-jETI allows automatic composition of functional units into software systems according to higher-level specifications using EDAM:
The iHOP Web service is annotated with EDAM concepts, either directly or via its use of BioXSD:
The Web services provided by the Computational Biology Unit (CBU) of the University of Bergen and its affiliated Uni Computing are annotated with EDAM concepts:
The eSysbio workbench for sharing and analysing bioinformatics data using public or private Web services and R scripts. eSysbio uses EDAM to annotate and denote the type and format of data items submitted to the system.
The SEQanswers wiki is an open catalogue of bioinformatics software tools, non-exclusively focussed on sequencing data analysis. SEQanswers tool wiki uses EDAM for annotation of the listed tools where applicable.
Last update: 2013-July-01